ABSTRACT
Background: Infection on surgical sites has become a major concern in the field of healthcare. Infection on the surgical site may occur up to 30 days from surgery and up to one year in patients receiving implants. Methods like excellent sterilisation techniques, surgical procedures, and availability of prophylactic antimicrobials are foremost needed. The present study was focused to assess bacteriological analysis and antibiotics consumption in various types of surgeries. Materials and Methods: An observational prospective study was conducted for three months in a tertiary care hospital. All subjects undergoing either clean, clean–contaminated, or contaminated surgeries admitted to the inpatient surgery, and gynaecology department were recruited. Microbial culture and antibiogram results of surgical site infection were recorded and analysed after IEC approval. Results: In this study, around one-third (50 patients; 31.25%), out of 160 patients were found with surgical site infection (SSI). The prevalence of SSI in females as compared to males was high. Furthermore, patients who had undergone clean–contaminated surgery were more prevalent in surgical site infections rather than patients undergoing other types of surgeries. The most frequently prescribed antibiotics by physicians were metronidazole, gentamycin, amikacin, ceftriaxone, and piperacillin with tazobactam. The most predominant pathogens were E. coli and Staphylococcus aureus at SSI. Moreover, E. coli, Staphylococcus aureus, Klebsiella pneumoniae, and Acinetobacter baumannii were resistant to most of the prescribed antibiotics but highly sensitive against ertapenem, imipenem, amikacin, gentamicin, tigecycline, colistin, linezolid, vancomycin, and teicoplanin. Conclusion: Gram-negative bacteria (E. coli) were the most common isolates associated with post-operative SSIs, followed by Staphylococcus aureus a gram-positive bacillus. Observation of high antibiotic resistance in the current study suggests the necessity for routine microbiological investigation of samples and their antibiogram in order to break the disease spread cycle of resistant microbes.
INTRODUCTION
Human skin consists of three-layer that is. epidermis, dermis, and endodermis, which all together act as a barrier in preventing foreign particles (bacteria, viruses, fungi, parasites) from invading into the human body. During most of the surgical procedures, this skin barrier is compromised in order to access, explore, repair, remove or transplant the desired internal organ, and the internal body gets directly exposed to a number of micro-organisms normally present in the environment.1 Infections acquired from operation theatres, other patients, hospital staff, and hospital facilities used in medical procedures are known as Hospital Acquired Infections (HAI). This type of infection is very hard to treat and in serious or worst cases can also be deadly because microorganisms in hospital facilities are most likely to be resistant to number of antibiotics.2 Every year an estimated 648,000 people in the U.S. develop infections during a hospital stay, and about 75,000 die, according to the Centres for Disease Control and Prevention (CDC). Out of every 100 hospitalised patients, seven patients in advanced countries, and ten patients in emerging countries acquire an HAI.3
Surgical Site Infections (SSI) are persistent and preventable health care associated infections. The major clinical manifestations of SSI are fever, redness, swelling, tenderness, warmth, pus drainage, and red streaks leading away from the wound.4 Prevention of post-operative infection, by all means is nowadays considered of paramount significance because it is impossible to create a predictably sterile wound. Despite improvements in prevention, SSIs remain a significant clinical problem as they are associated with substantial mortality/morbidity, increased length of stay in the hospital, increased cost of treatment, and impose severe demands on healthcare resources.3 Approximately 25% of patients with SSIs develop severe sepsis and shock and are moved to an ICU. According to CDC data for 2018, the morbidity was 157,500 for surgical site infections (SSI) in the US, with an estimated mortality of 8,205. It is a burden for the patient with an additional 11 days of hospitalisation for each SSI and a burden to the system with an overall cost of $3.2 billion per year.5 Surgical site infections are having high incidence rate of 20%, which may vary depending on the type of surgical procedures and 11% of all deaths in intensive care units were associated with SSI. Surgical wound contamination potentials, patients’ clinical conditions, type of surgery, and length of surgery were variables statistically significantly associated with SSIs, and should be viewed as risk factors.6 Most commonly isolated pathogens responsible for causing surgical site infections are Staphylococcus aureus, coagulase negative Staphylococci, Enterococcus spp. and Escherichia coli.7 In present scenario, it is challenging to provide accurate data for postoperative wound infections in term of variety of specialties, operations, patients, and geographies. Also, identifying wound infections is more challenging due to the increased prevalence of day case surgery, and shortened hospital stays.8 To improve patient healthcare, a practise of surveillance after critical operative procedures, to overcome SSI in patient of wards, or ICU. In this view, the present study was conducted to access the schedule of antibiotics as prophylaxis in different types of surgeries, and to identify various microbes involved in causing surgical site infection. This study will help physicians to utilise this data in prescribing antibiotics as prophylaxis more efficiently according to various microbes, which are most abundantly causing infections in health care facilities.
MATERIALS AND METHODS
Study Design
An observational study on antibiotic consumption and bacteriological analysis was conducted by the department of Pharmacy Practise in collaboration with surgery department of M.M. Institute of Medical Sciences Hospital, with a sample size of 90 patients. The study was conducted for a total time period of 6 months from October 2021 to March 2021. The Protocol of this study and case report form was approved by institutional ethics committee. MMDU|MMIMSR|IEC|1864. Detailed objectives of the study were explained to the patients and their guardians who meet the inclusion criteria. Informed consent form was taken from the patient (or guardian) willing to participate in the study. Patient selection was done on the basis of inclusion criteria. Subjects were informed that withdrawal from the study or denial to participate will not lead to loss of benefits and penalty.
Study Tools
- Patients who meet the inclusion criteria.
- Age >18 years.
- Both male and female patients.
- Clean, clean contaminated, and contaminated surgeries only.
- Must have antibiotic sensitivity report and microbiological culture done.
- Confirmed case of surgical site infection presence.
- Patients underwent elective surgical procedures.
Their data was collected on the case report form (CRF). It included patient’s demographics, surgery notes, antibiotic prescription chat, microbiological culture test, and antibiotic sensitivity test.
Method of Sample Collection in Case of SSI
Sample was collected before starting antimicrobial therapy. Specimen was collected by a qualified nurse. Samples of pus were preferred as swabs. If possible, a few ml of pus in a sterile universal bottle, or even a few drops still in a syringe was preferred than a swab sample. Frequency and numbers of specimen collection were dependent on clinical condition of the patient until the patient is treated. Ideally, a minimum volume of 1 ml pus sample was required. The volume of specimen influences the transport time that is acceptable. Large volumes of purulent material maintain the viability of anaerobes for longer duration.
Procedure of Pus Culture and Antibiogram
Isolation, then identification of the microbe, and antibiogram of a pathogenic agent were normally carried out when a bacterial disease has complications like septicemia. Following diffusion of the compounds through the agar, a “halo” or zone of inhibition forms where a concentration of the specific diffused antibiotic was sufficient to inhibit that microbial growth. The correct explanation of the antibiogram was of interest to microbiologists and laboratory technicians alike. Standardised methods were established and could be found in the WHO manuals.
The collected data were tabulated in Microsoft Office Excel software. Data collected were presented using tables, charts, and bar graphs
RESULTS
This is an observational study conducted in order to assess the schedule of prophylactic antibiotics in different type of surgery, utilisation pattern of antibiotics post-surgery, and microbiological analysis of bacteria in post-operative infections.
From demographic details of patient’s data showed 76.67% of females and 23.34% males contracted infection of surgical site. This data suggests that the population of females are at higher risk of contracting SSI. Most number of patients acquired SSI were from age group of 38–47 years followed by age group of 28–37, 18–27, and 48–57 years (Table 1). From the total, 77% patients contracted SSI underwent clean–contaminated wound classification/surgery and 13% patients of with contaminated surgical procedures contracted SSI. Patients undergone clean surgical procedures were 10%.
| Age Group | No. of Patients | |
|---|---|---|
| Male | Female | |
| 18–27 | 3 | 12 |
| 28–37 | 6 | 15 |
| 38–47 | 6 | 24 |
| 48–57 | 3 | 9 |
| 58–67 | 3 | 6 |
| 68–77 | 0 | 3 |
| TOTAL | 21 | 69 |
| Total Percentage | 23.34% | 76.67% |
Post-operative antibiotics prescribed were metronidazole (27%) was the most prescribed post-operative antibiotic followed by amikacin with gentamycin (12%), piperacillin with tazobactam (10%), ceftriaxone (7%), ciprofloxacin, ceftriaxone with sulbactam with 5%, meropenem, clindamycin, linezolid (4%), antibiotics like colistin, cefuroxime, ampicillin, cefepime with sulbactam, amoxicillin with clavulanic acid at only 2% and cefixime and tigecycline were prescribed at rate of 1% post-operation (Figure 1).
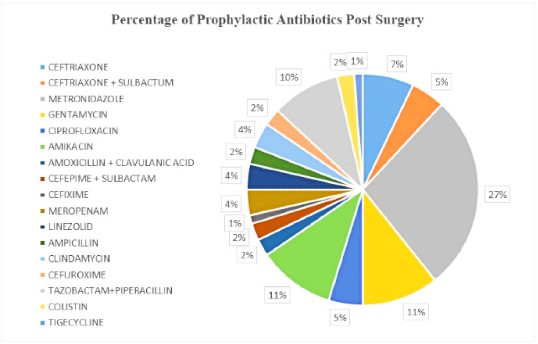
Figure 1.
Distribution of prophylactic antibiotics Prescribed Post-Surgery.
The antibiotics prescribed after clean surgery were ciprofloxacin (33.3%), amikacin (66.6%), tazobactam with piperacillin (66.6%), tigecycline (33.3%) as BD; metronidazole (100%), meropenem (33.3%), colistin 33.3% as TDS In clean–contaminated surgery antibiotics prescribed post-surgery were amikacin (4.3%) as OD; ceftriaxone with sulbactam (13%), ceftriaxone (26%), ciprofloxacin (8.6%), amikacin (21%), amoxicillin with clavulanic acid (8.6%), cefepime with sulbactam (8.6%), cefixime (4.3%), linezolid (4.3%), cefuroxime (8.6%) and piperacillin with tazobactam (8.6%) as BD; metronidazole (73.9%), gentamycin (34.7%), amikacin (4.3%), meropenem (4.3%), ampicillin (4.3%), clindamycin (8.6%) and piperacillin with tazobactam (8.6%) as TDS; piperacillin with tazobactam (4.3%) as QID. In contaminated surgery antibiotics used post-surgery were linezolid (25%) as OD; ceftriaxone with sulbactam (25%), gentamycin (25%), ciprofloxacin (25%), amikacin (25%), linezolid (25%), clindamycin (25%) as BD; metronidazole (75%), meropenem (25%), ampicillin (25%), piperacillin with tazobactam (25%) as TDS (Table 2).
| Type of Surgery/ Frequency | OD | BD | TDS | QID |
|---|---|---|---|---|
| Clean (9) | – | Ciprofloxacin (33.3%),Amikacin (66.6%),Piperacillin+Tazobactam (66.6%),Tigecycline (33.3%) | Metronidazole (100%),Meropenem (33.3%), Colistin (33.3%) | – |
| Clean Contaminated (69) | Amikacin (4.3%), | Ceftriaxone + Sulbactam (13%), Ceftriaxone (26%),Ciprofloxacin (8.6%), Amikacin (21%), Amoxicillin + Clavulanic Acid (8.6%), Cefepime + Sulbactam (8.6%),Cefixime (4.3%), Linezolid (4.3%), Cefuroxime (8.6%), Piperacillin+Tazobactam (8.6%) | Metronidazole (73.9%), Gentamycin (34.7), Amikacin (4.3%), Meropenam (4.3%), Ampicillin (4.3%), Clindamycin (8.6%), Piperacillin+Tazobactam (8.6%), | Piperacillin+Tazobactam (4.3%) |
| Contaminated (12) | Linezolid (25%) | Ceftriaxone + Sulbactam (25%), Gentamycin (25%), Ciprofloxacin (25%), Amikacin (25%), Linezolid (25%), Clindamycin (25%) | Metronidazole (75%), Meropenem (25%),Ampicillin (25%), Piperacillin+Tazobactam (25%), | – |
Data represent the microbes that caused SSI on a clean wound post-operation. Microbes such as E. coli, Klebsiella pneumoniae, Acinetobacter baumannii were found in equal proportions (Figure 2 A). Microbes that caused post-operative SSI in clean-contaminated surgery were: – E. coli (31%), Staphylococcus aureus (26%), Klebsiella pneumoniae (17%), Enterococcus species (13%), Pseudomonas aeruginosa (9%) and Acinetobacter baumannii (4%) (Figure 2B). Microbes caused post-operative SSI in contaminated wounds/surgery were E. coli (75%) and Staphylococcus aureus (25%) (Figure 2 C).
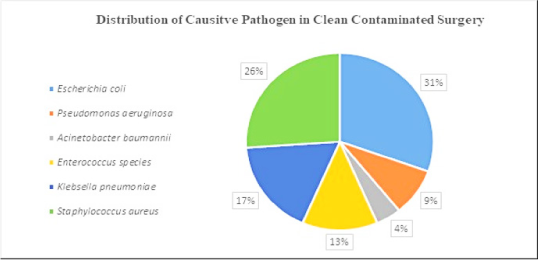
Figure 2 A.
Distribution of causative pathogen in clean surgery.
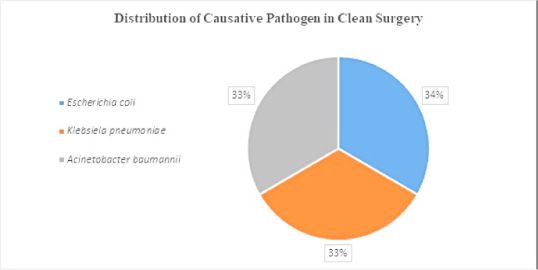
Figure 2 B.
Distribution of causative pathogen in clean contaminated surgery.
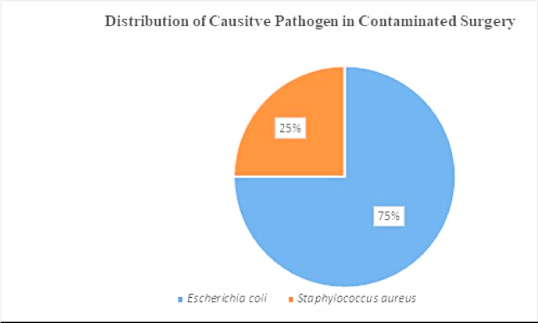
Figure 2 C.
Distribution of causative pathogen in contaminated surgery.
Sensitivity and Resistant pattern of antibiotics with microbes
Microbes which were isolated from the surgical site shown in Figure 3, were incubated for 48hrs, and tested against different antibiotics. Microbes isolated were E. coli, Staphylococcus aureus, Klebsiella pneumoniae, Enterococcus spp., Acinetobacter baumannii. In this study, E. coli was found to be resistant against amoxicillin, cefuroxime, cefotaxime, ceftriaxone, ciprofloxacin, trimethoprim, piperacillin with tazobactam, imipenem, amikacin, gentamicin, cefepime, meropenem, ampicillin, tobramycin, netilmicin, and the least to amoxicillin with clavulanic acid. Microbe such as Staphylococcus aureus was found to be having resistance against ciprofloxacin, trimethoprim, gentamicin, levofloxacin, erythromycin, tetracycline, vancomycin, penicillin, clindamycin, oxacillin, cefoxitin. Microbe such as Klebsiella pneumoniae was found to be having resistance against cefuroxime, cefotaxime, ceftriaxone, ciprofloxacin, trimethoprim, piperacillin with tazobactam, imipenem, amikacin, gentamicin, cefepime, meropenem, and amoxicillin with clavulanic acid. Microbe like Enterococcus spp. was found that it was having resistance against ciprofloxacin, gentamicin, levofloxacin, erythromycin, tetracycline, fosfomycin, and norfloxacin, ceftazidime, levofloxacin, doripenem, ticarcillin with clavulanic acid, cefepime, meropenem, tobramycin, and netilmicin. Microbes such as Acinetobacter baumannii were found to be having resistance against ciprofloxacin, trimethoprim, piperacillin with tazobactam, imipenem, amikacin, gentamicin (Figure 4).
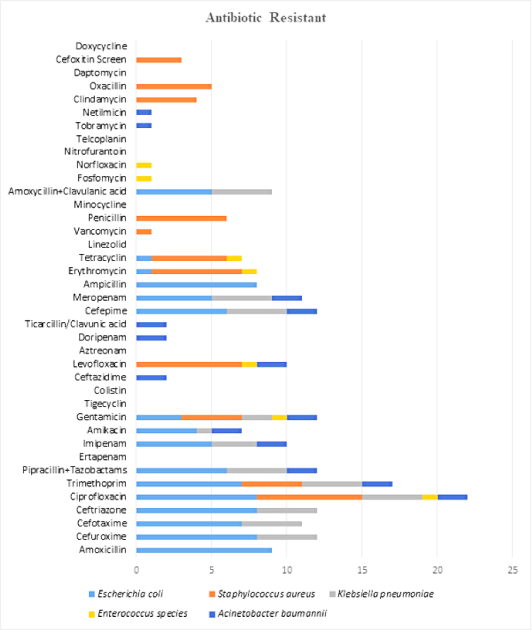
Figure 3.
Graph of Resistance Pattern of Different Microbes to Different Antibiotics.
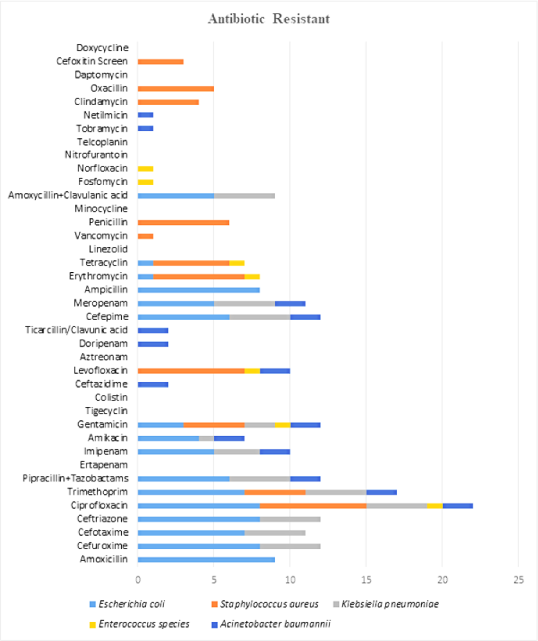
Figure 4.
Graph of Resistance Pattern of Different Microbes to Different Antibiotics.
The pattern of sensitivity of different microbes towards different antibiotics shows in Figure. Microbe such as E. coli were found to be sensitive towards trimethoprim, piperacillin with tazobactam, ertapenem, imipenem, amikacin, gentamicin, tigecycline, colistin, cefepime, meropenem, linezolid, vancomycin, and amoxicillin with clavulanic acid. Microbes such as Staphylococcus aureus was having sensitivity towards trimethoprim, gentamicin, erythromycin, tetracycline, linezolid, vancomycin, minocycline, teicoplanin, clindamycin, daptomycin, and doxycycline. Microbes such as Klebsiella pneumoniae was having sensitivity towards antibiotics ciprofloxacin, amikacin, gentamicin, tigecycline, and colistin. Microbe such as Enterococcus spp. was sensitive towards antibiotics amoxicillin, piperacillin with tazobactam, ticarcillin with clavulanic acid, ampicillin, linezolid, vancomycin, penicillin, amoxicillin with clavulanic acid, nitrofurantoin, and teicoplanin. Microbe such as Acinetobacter baumannii was sensitive towards antibiotics tigecycline and colistin (Figure 4).
DISCUSSION
This observational study was conducted over a period of six months and helped to record immensely important findings. In this study, predominantly female population was found to be more prone to acquire Surgical Site Infections (SSIs). A study by dessie et al. has shown that majorly women population were getting infected predominantly by 52.3% from SSIs.9 In the present study, most of patients got infected from SSIs were from age group of 42.5 yrs. Similar results by Goswami et al. that shows mean age of patients getting infected from SSI were 37.4 years. SSIs in clean-contaminated wounds/surgery were in most of cases in the present study.10 A study by Mundhada et al. has shown 39.39% who got post-operative SSI were of clean and contaminated wound. This seems a little ironical because contaminated and dirty surgery may involve non-sterile methods.11 This is due to ignorance of post-operative care like in proper cleaning and sterile dressing of wound or due to improper sterilisation process of equipment’s.11 In this study, the antibiotic prescribed to the greatest number of patient population was metronidazole 27% which is a narrow spectrum antimicrobial drug and covers most of the anaerobes. The result was similar with a study conducted by Misganaw et al. showed that, out of total 68 SSI patients, the prescribed antibiotics were in a combination of ceftriaxone, and metronidazole 47.46%.12 Moreover, antibiotics like ceftriaxone, amikacin, and gentamycin were also prescribed post-surgery in this study. Microbes isolated from the surgical site were E. coli, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Enterococcus spp., Pseudomonas aeruginosa in different type of clean, clean–contaminated, and contaminated wound. A study conducted by Vardhana et al. in India shows that microbes isolated predominantly were Staphylococcus aureus (44.8%), E. coli (42%), Methicillin–resistant Staphylococcus aureus (31%), Klebsiella spp. (29.8%), Pseudomonas aeruginosa (26.3%), CoNS, and Enterococcus spp. (10.3%).13 Getting resistant to antibiotics slowly is a complete terrifying scenario but getting infected by microbes which are already resistant to a lot of widely used broad and narrow spectrum antibiotics can be fatal. Similar national study conducted by Aniruddha et al. was also conducted, who underwent clean, and clean–contaminated surgery, Shows the most prevalent pathogen identified was streptococcus aureus followed by E. coli, P. aeruginosa, K. pneumoniae, Acinetobacter spp. and S. epidermis. It was discovered that sensitivity of these microbes towards some antibiotics like ciprofloxacin, ampicillin, cefotaxime, tetracycline.14 In our study, microbe isolated largely from surgical site was E. coli and found to be resistant among various antibiotics such as amoxicillin, cefuroxime, cefotaxime, ceftriaxone, ciprofloxacin, trimethoprim, piperacillin with tazobactam, imipenem, amikacin, gentamycin, cefepime, meropenem, ampicillin, erythromycin, tetracycline, amoxicillin with clavulanic acid (varied from individual–to-individual case). A study conducted by Ali et al. showed that E. coli was the second predominant microbe on site of SSI and was resistant to various antibiotics such as cefoxitin, cefepime, cephalothin, ceftazidime, amoxicillin with clavulanic acid, ciprofloxacin, ceftriaxone, gentamicin, azithromycin, trimethoprim with sulfamethoxazole.15 Along with sensitivity study of different microbes towards different antibiotics our study also focused on determination of SSI prevalence in different age–group, gender, type of surgery/ wound, and different microbes in different departments. Main objective of our study is to aid in infection control and prevent emergence of resistant microbes. There were some limitations due to COVID-19 which led to reduced sample size as less people underwent elective surgery.
CONCLUSION
In conclusion, gram negative bacteria (E. coli) were the most common isolates associated with post-operative SSIs, followed by Staphylococcus aureus a gram-positive bacillus. Moreover, isolated microbes from SSI patients were resistant to most of the antibiotics used. Prevalence of these anaerobes in the surgery wards (hospital environment) and surgical instruments used by health professionals are most common cause of SSI rather than patients themselves. Observation of high antibiotics resistance in current study suggest the necessity for routine microbiological investigation of samples and their antibiogram. Additionally, physician prescriptions should be written according to current updates on the antibiotic, and results of patient medical profiles so that healthcare professionals can prevent the emergence of microbe resistance to break disease spread cycle.
References
- Young B, O’Dowd G, Woodford P, Wheater P. Wheater’s Functional histology [cited Aug 10, 2021]. 2014 eBook ISBN: 9780702054884 [Google Scholar]
- Monegro AF, Muppidi V, Regunath H. Hospital acquired infections. StatPearls [Internet]. Updated 2022:2022 [Google Scholar]
- Haque M, Sartelli M, McKimm J, Abu Bakar M. Health care-associated infections – an overview. Infect Drug Resist. 2018;11:2321-33. [PubMed] | [CrossRef] | [Google Scholar]
- Swenson BR, Hedrick TL, Metzger R, Bonatti H, Pruett TL, Sawyer RG, et al. Effects of preoperative skin preparation on postoperative wound infection rates: A prospective study of 3 skin preparation protocols. Infect Control Hosp Epidemiol. 2009;30(10):964-71. [PubMed] | [CrossRef] | [Google Scholar]
- Zabaglo M, Sharman T. Postoperative wound infection. StatPearls [Internet]. Updated 2021:2022 [PubMed] | [CrossRef] | [Google Scholar]
- Atilla A, Doğanay Z, Kefeli Çelik H, Demirağ MD, S Kiliç S. Central line-associated blood stream infections: Characteristics and risk factors for mortality over a 5.5-year period. Turk J Med Sci. 2017;47(2):646-52. [PubMed] | [CrossRef] | [Google Scholar]
- Alemkere G. Antibiotic usage in surgical prophylaxis: A prospective observational study in the surgical ward of Nekemte referral hospital. PLOS ONE. 2018;13(9):e0203523 [PubMed] | [CrossRef] | [Google Scholar]
- Spagnolo AM, Ottria G, Amicizia D, Perdelli F, Cristina ML. Operating theatre quality and prevention of surgical site Infections. J Prev Med Hyg. 2013;54(3):131-7. [PubMed] | [Google Scholar]
- Dessie W, Mulugeta G, Fentaw S, Mihret A, Hassen M, Abebe E, et al. Pattern of bacterial pathogens and their susceptibility isolated from surgical site infections at selected referral hospitals, Addis Ababa, Ethiopia. Int J Microbiol. 2016;2016:2418902 [PubMed] | [CrossRef] | [Google Scholar]
- Goswami NN, Trivedi HR, Goswami AP, Patel TK, Tripathi CB. Antibiotic sensitivity profile of bacterial pathogens in postoperative wound infections at a tertiary care hospital in Gujarat, India. J Pharmacol Pharmacother. 2011;2(3):158-64. [PubMed] | [CrossRef] | [Google Scholar]
- Mundhada AS, Tenpe S. A study of organisms causing surgical site infections and their antimicrobial susceptibility in a tertiary care government hospital. Indian J Pathol Microbiol. 2015;58(2):195-200. [PubMed] | [CrossRef] | [Google Scholar]
- Misganaw D, Linger B, Abesha A. Surgical antibiotic prophylaxis use and surgical site infection pattern in Dessie referral hospital, Dessie, Northeast of Ethiopia. BioMed Res Int. 2020:1695683 [PubMed] | [CrossRef] | [Google Scholar]
- Vishnu Vardhana Rao KV, Pradeep MSS. A Study on surgical Site Infections, their bacteriological profile and antimicrobial susceptibility pattern. IJMMTD. 2019;5(1):9-13. [CrossRef] | [Google Scholar]
- Pal S, Sayana A, Joshi A, Juyal D. Array. J Family Med Prim Care. Array;8(11):3600-6. [PubMed] | [CrossRef] | [Google Scholar]
- Ali KM, Al-Jaff BMA. Source and antibiotic susceptibility of gram-negative bacteria causing superficial incisional surgical site infections. Int J Surg Open. 2021;30:100318 [CrossRef] | [Google Scholar]
