ABSTRACT
The drug discovery process has been a challenging task for any pharmaceutical industry. In ancient times the discovery of new drug molecules was made on an experimental basis until computers were introduced in the field of Pharmaceutical sciences, which led to a breakthrough in pharmaceutical research and development. Computer-Aided Drug Design (CADD) revolutionized the drug discovery process by minimizing the time and showing accurate results in the drug discovery process. Artificial Intelligence (AI) and Machine Learning (ML) are the advanced version of CADD technologies for the drug discovery process. The integrated technologies of both AI and machine learning have unleashed the immense potential of researchers in the pharmaceutical industries and drug discovery process, leading to significant innovations and the development of intelligent technologies. This review highlights some of them as it focuses on the concept of industrialization 4.0 (smart industries), which involves CADD’s most progressive and innovative ideas like automation, augmented reality, digital twins, cobots, and computational fluid dynamics. These are the advanced form of CADD technologies, which have immense potential for designing smart industries. All these have led the pharmaceutical industries to evolve from man to machine, which results in complete automation.
INTRODUCTION
Artificial Intelligence (AI) is defined as the ability of the system to interpret, learn from external data sources, and produce accurate and desired results.1 The concept of AI is spread throughout various fields of Information Technology (IT) sectors, automation, supercomputers, robotics, pharmaceutical sectors, etc. The idea of AI is prevalent in the IT and engineering sector (i.e., from your smartphones to self-driving Tesla cars). In the medical industry, robotic surgeries and other advanced AI technologies have highlighted the immense potential of artificial intelligence worldwide. Our pharmaceutical industries are also taking part in the field of automation, so they can also build colossal profit and enhance their production rates in the world of medicines. Most of them have indulged automated machines and software in their companies to produce faster results in the production of drugs, i.e., the world as a whole is moving towards automation with the help of AI. The concept of AI in Computer-Aided Drug Design (CADD) has changed the working environment of pharmaceutical companies. There was a time when around 15-20 years were taken to discover new drug molecules, but due to AI and automation, the period for finding new molecules is reduced to 5-6 months. Ease of work and complicated and unnecessary steps involved in drug discovery were eliminated.2
The concept of AI can be applied to Structure-Based Drug Design (SBDD) and Ligand-Based Drug Design (LBDD) concepts. Machine Learning and AI play a crucial role in CADD. Both of them have an equivalent position in computer-aided drug design. The most fundamental part of AI is Artificial Neural Network (ANN), also referred to as the “Digitalized Human Brain Model.” ANN plays a very significant role in the molecular modeling of pharmaceutical sciences.3 The application of ANN is very well established in various sectors of computer-aided drug design, and it has been summarized in Figure 5. An ideal AI software should possess the following properties.4
Learn from experience
- Recognize and understand images.
- Solve complex algorithmic problems.
- Understand and produce desired solutions.
HISTORICAL ASPECTS OF ARTIFICIAL INTELLIGENCE
The seeds of AI have already been sowed during the ancient Greek period. It was believed that the concept of humanoid robots started during this period. For example, at that time, it is thought that a wind mythologist named Daedalus ruled the mythology of wind and tried to create artificial humans.5 The concept of modern AI had its beginning in the 1940s. It all started in 1942 when for the first time, a short science fiction story got published by an American science fiction author. The story was about the two scientists who developed the concept of robotics, but this theory was a frictional concept coming from the frictional book.6 But during the Second World War, British mathematician Alan Turing developed a decoding machine known as Bombe for decoding a secret code sent by Germens, which was very helpful for the British government. Bombe was a giant electro-mechanical computer that successfully cracked. After his great innovation, Alan couldn’t believe it and started to wonder how a machine could be so intelligent; in 1950, Alan published an article called “Computing Machinery and Intelligence”. His report explained how to develop and test your intelligent machine; this made a massive breakthrough in the arena of technologies. His article became the basic fundaments for advanced robotics and computer science.7 He was known as the father of computer science.
J McCarthy is known as the father of computer science. He and Marvin Minsky coined the word “Artificial Intelligence” in 1956. They hosted the first summer AI workshop named Dartmouth Summer Research Project on AI (DSRPAI), where most participants were computer scientists; among them, one was N. Rochester, who developed the first scientific computer, IBM 701.8
Other essential innovations took place during different periods, such as the famous computer program ELIZA, created by Weizenbaum from 1964-to 1966. This program was the first natural language stimulation program that could talk with humans.9 IBM developed its “deep blue” program in 1997, and this program was able to beat professional chess players. The program used the tree search method to estimate 200 million possible moves and was 20 moves far ahead than usual in chess games.10 Statistical AI models were developed in the early 1940s by Canadian psychologist Donald Olding Hebb, a learning theory for machines known as Hebbian Learning, which was considered to be an exact replicate of human neurons, leading to the discovery of Artificial Neural networks.11 The work on ANN started in 2015. The first glimpse of ANN’s work was the development of the Alpha Go program by Google. The ANN algorithm used in Alpha Go was deep learning.12 ANN, deep learning, and machine learning are the fundamentals of AI, and these concepts are applicable in various arrears. Examples include the image recognition algorithm used by Facebook and the speech reorganizing algorithm in smart speakers like Alexa and self-driving cars like Tesla.13 The entire timeline of the history of Artificial Intelligence is summarized in chronological order and shown in Figure 1.
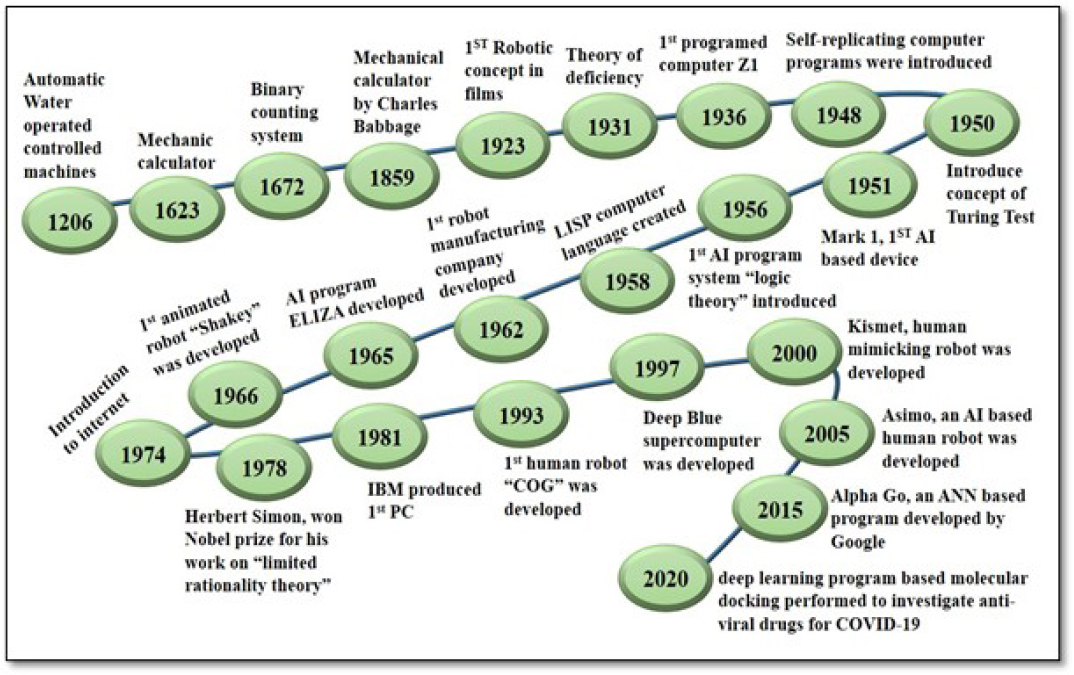
Figure 1:
Structural Outline of History of Artificial Intelligence.15
ARTIFICIAL NEURAL NETWORK
ANN are also known as the “Digitalized Human Brain.” It consists of three significant compartments (brain compartment): input compartment, hidden compartment, and output compartment. These three compartments are connected by neural networks (brain neurons).14–16 These neural networks (Neurons) are complex algorithm-based networks sequenced regularly in-between these compartments and help transfer the information between these compartments. The Input compartment, where we feed our input data, is connected with the hidden compartment, where data analysis takes place. According to that, it will back-propagate (denies the input data) or forward the data toward the output compartment as a result of input data; as shown in Figure 2, it is believed that the molecular modeling and drug design are highly dependent on the concepts of ANN. It reduces the complexity of the statistical modeling used in HTVS, QSAR modeling, pharmacokinetics, and pharmacodynamics studies involved in drug discovery. It has an outstanding interpreting ability, which interprets linear and non-linear computational parameters and helps drug discovery.2,17
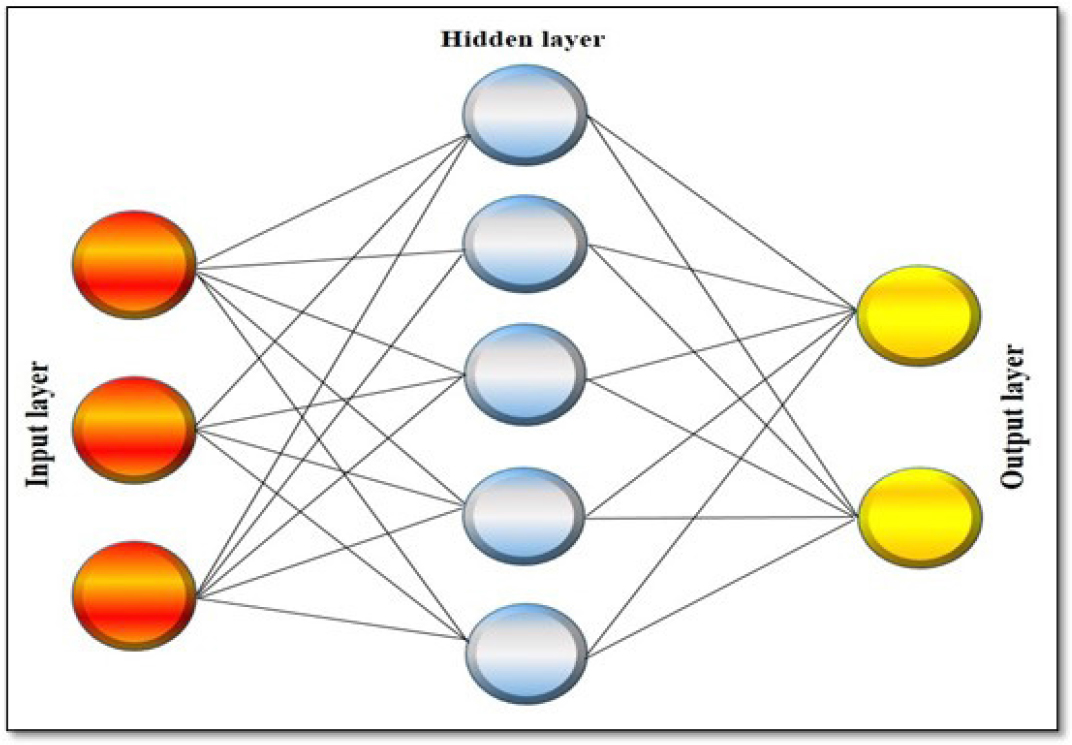
Figure 2:
Diagrammatic representation of Artificial Neural Network.20
DEEP LEARNING
DL neural networks possess an Input, multiple hidden, and an output compartment, as shown in Figure 3; it can learn most complex modules containing adequate data given by different neurons and input sets and provide us with highly accurate outputs. The most commonly applied processing methods are the backed-propagation algorithm and gradient-based optimization techniques along with other novel methods like Initialization schemes and neural activation functions have been beneficial in training deep learning networks.2,18 The processing techniques are of three types, Recurrent Neural Network (RNN), Conventional Neural Network (CNN), and Graph Neural Network (GNN).
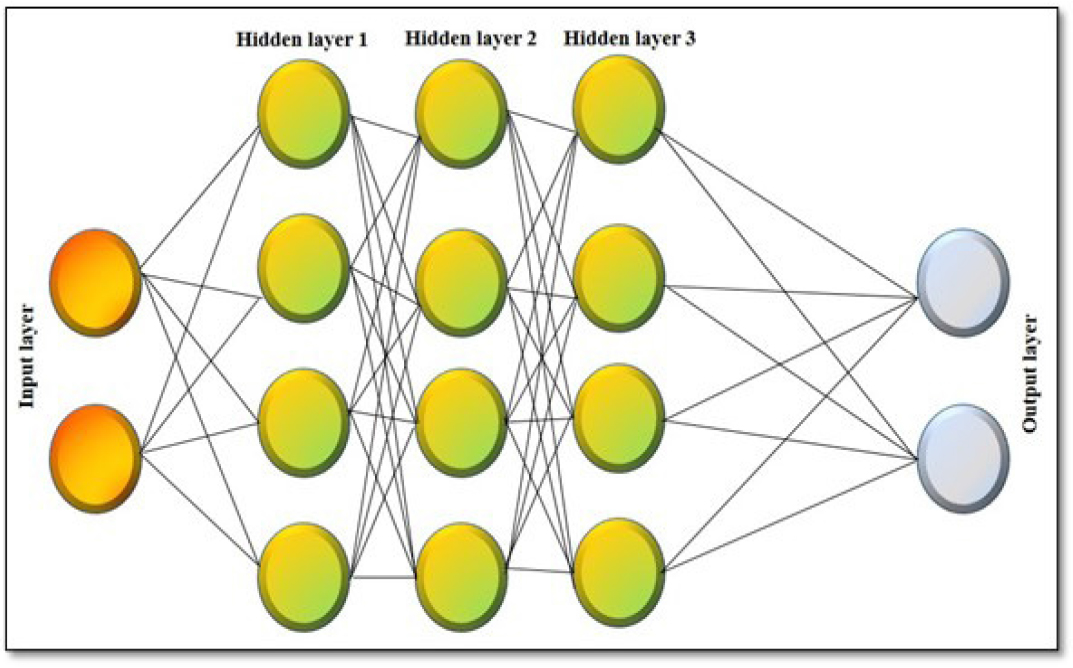
Figure 3:
Schematic diagram of Deep learning.23
Recurrent Neural Network – RNN algorithms are developed to read and analyze sequence-level data. It works based on the internal memory principle, just as we humans have our subconscious mind, which acts as a reservoir of memories; similarly, it acts as a reservoir of memories for any machine learning program. It was discovered very early, around 1980 (the golden period of computational chemistry). Still, only now, in recent years, can we understand and apply its potential in various aspects of fields. The application can be found in Google’s voice assistance, APPLE’s SIRI voice command, and other AI-based voice recognition programs.2 A schematic representation of CNN is shown in Figure 4.
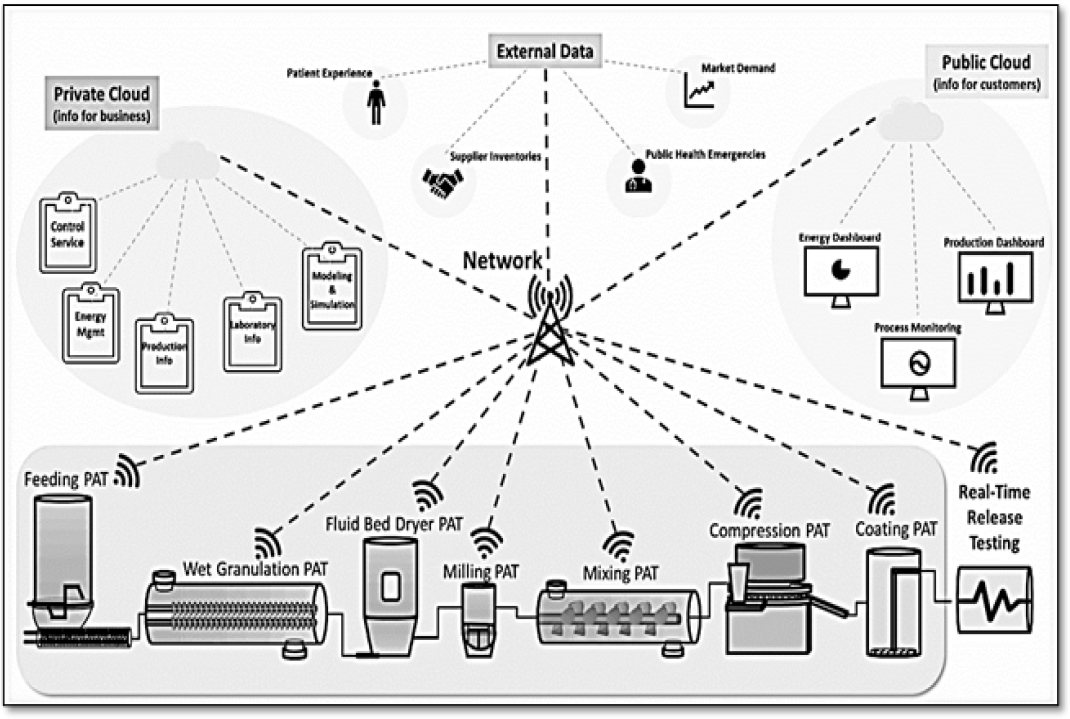
Figure 4:
Schematic representation of different types of deep learning, were (a).represents RNN, (b).represents CNN, (c).represents GNN.30
Conventional Neural Network – It is used mainly to analyze images. It’s a visual analyzer. It was first developed during the 1980’s period. CNN consists of multiple hidden layers and specific filter layers, which help them analyze the pixels of images and produce desired output results.2,19,20 A schematic representation of CNN is shown in Figure 4.
Graph Neural Network – To understand GNN, first, we need to under the meaning of a graph in AI Graph refers to the set of un-organized data and its connections between them for example, images, texts, molecules, social networks, etc. Scientists have utilized this information to develop the GNN algorithm. It consists of two components, vertices and envelopes.21–23 It can be applied in text classification, neural machine translation, object detection, molecular fingerprints, etc.,24 A schematic representation of GNN is represented in Figure 4.
MACHINE LEARNING
ML is a basic model with multiple algorithms to identify the pattern within data. Deep learning comes under machine learning.25 It acts precisely as the human brain neurons by transmitting information via electronic impulses. Their complex algorithms, which are nothing but mathematical models, use underlying algorithms in the given data and information to learn and predict future data. It can solve various complex mathematical problems in the data and produce accurate results. It is broadly categorized into supervised and un-supervised learning models.
Supervised learning- the mathematical model represents the data relationship in the form of input and output variables. Although, it is challenging to understand the linear relation between them.26
Un-supervised learning- these learning models hunt for the hidden patterns present within the input data, analyze them and analyze the relationship between the data points.24
Reinforcement learning models are similar to supervised models. Still, the only difference is that they mainly deal with complex problems and learn to reproduce a series of actions while figuring out those problems, to avoid unwanted information related to the problem, and produce similar output just as supervised learning.27,28
APPLICATION OF MACHINE LEARNING IN DRUG DISCOVERY PROCESS
Target Identification – the ML method was used to develop a model to overcome the challenges related to DNA-protein pathways and process them for a genetic level.27 The Support Vector Machine (SVM) method was developed to analyze the genomic sequencing to distinguish between protein-based homologs in the case of breast, pancreatic, and ovarian cancers.29 SVM is also used to identify the drug target by utilizing the biomarker approach in muscle tissue to determine the drug targets for treating the aging problem of human tissue.30,31
High-Throughput Virtual Screening – Using different SVM methods like (Naïve Bayes, Spy, and Rocchio) around 3000 targets were predicted and ranked; these targets were associated with the aging gene, which was used to treat the aging problem in humans.32 SVM technique was also used for G-protein coupled receptors, screening out 129,994 compounds; from these, only 1573 compounds were short-listed, which has a very high affinity towards G-protein coupled receptors. This method has a 69% higher efficiency than the standard virtual screening.33
Structure-Based Drug Design – Self-Organizing Maps based Prediction of the Drug Equivalence Relationship (SPiDER) is mainly used in De-novo drug design to identify the new chemical structure fitting the pharmacological model’s space for small molecules.34 Deep-Chem is n free source tool used to construct, provide and evaluate complex models and induce ligand screening for commercial drugs.35,36 DeepTox is another multi-task ANN tool used to calculate chemical descriptors and trains ANN to determine nuclear toxicity by normalizing chemical structure. This method was implicated in determining the toxicity of small molecules through high-throughput virtual screenings.37
Protein-Protein Interaction Prediction – STRING software is one of the widely used Protein-Protein Interaction (PPI) databases, consisting of 1.4 billion PPIs; the software interface predicts the protein-protein binding sites composed of amino acids.36 Software like Z-DOCK and Symm-Dock is a protein-protein docking software that uses SVM and NBS principles to predict protein-protein interaction.38 Another method, Fragment Docking-Direct Coupling analysis (FD-DC), is used to predict drugs acting on protein-protein interaction sites. This method initially develops fragments using iFitDock software to predict the hotspot in the protein-protein interaction interface.39
Ligand-Based Drug Design – ML tools like Random Forest (RF), SVM, and ANN are most commonly used to design Quantitative Structure Activity Relationship (QSAR) models.40,41 In 2012, a state of art method was developed with multiple deep neural networks to create RF-based QSAR models.42
Prediction of Pharmacokinetics properties – For the prediction of ADME/T properties of drug molecules, several DL methods can be used. For example, the lead molecule’s solubility prediction was made using the Conventional Neural Network-Artificial Neural Network (CNN-ANN) algorithm.43 Another method, called Molecular Graph Encoding-Convolutional Neural Network (MGE-CNN), which was used to predict acute oral toxicity, produced more accurate results than the traditional method.44 DNN method was used to develop DeepTox software, which was used for toxicity prediction.45 Immobilized Artificial Membrane (IAM) methods were used to predict the quantitative characterization of lipophilicity.46
PHARMACEUTICAL INDUSTRIES TOWARDS AUTOMATION: INDUSTRIALIZATION 4.0
An idea to make pharmaceuticals a smart and labor-less industries. Industrialization 4.0 is an architect of AI, integrated connectivity, and robotics, where the entire system is accessible with one click of a button (i.e., zero involvement of humans).47 To optimize the whole manufacturing system, the concept of automation and the robotic system helps to integrate original-time and online data with the production process and artificial intelligence.48 To design an intelligent factory, the concept of Industrialization 4.0 possess key characteristic features, like industrial wireless connectivity, artificial intelligence, and the idea of full automation. Industrial wireless connectivity is an integrated cyber-based system, including computing devices, sensors, instruments, and equipment.
All these are interconnected online with each other in a cohesive network. The entire manual data must be digitalized, called data digitalization.49 For example, the pharmaceutical manufacturing process includes information about the supply chain process, the procedure, and the operator’s instructions.49 Real-time operations are monitored by using videos.50 A cyber-based pharmaceutical production unit’s critical parameters include private cloud, public cloud, and manufacturing ground. A private cloud (cloud-based computing device) gathers business-related information (control system, production information, lab information, modeling, and simulation), and a public cloud collects information about product management and process monitoring. Both digitally display the status of the entire physical system, enabling real-time prediction and optimization of the system. Manufacturing ground covers the equipment, Process Analytical Technology (PAT) instrumentation, and real-time release testing. PAT is a Quality-based Design (QbD) technology that mainly focuses on the quality of the product and controls the quality product profile. PAT maintains and monitors the manufacturing process, and real-time release testing assures the quality of the product during the ongoing manufacturing process.51 The entire cyber-based manufacturing unit is shown in Figure 5.
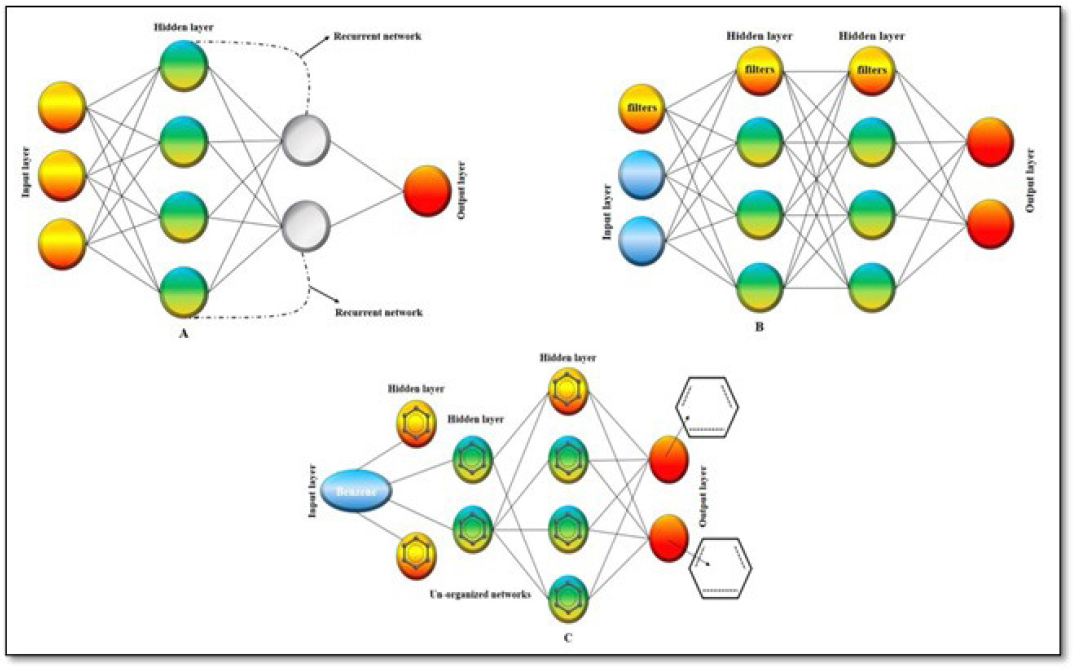
Figure 5:
Schematic representation of cyber-based manufacturing unit.67
It has solved significant problems by digitalizing the entire system, which ultimately reduced the man cost of industry and enhanced the efficiency and productivity of pharmaceutical industries. It also facilitates the understanding and visualization of digitalized manufacturing operations.
AI is considered the backbone of smart factories; ANN and ML have been part of AI, and their algorithms helped us establish smart factories. Al helps to create an overall digitalized and automated environment. It also reduces human interference and helps optimize and enhance the manufacturing process efficiency in pharmaceutical industries. Smart factories with advanced technologies like computerized visualization quality control, collaborative robots, digital twins, and real-time augmented reality are a few examples of AI technologies. Computerized visualization-based quality control, where AI software does the quality control test for packaging, labels, glass vials, and other pharmaceutical parameters.52 It uses the images to analyze the deviations occurring with the standard image.
Collaborative robots, also called “cobots,” are AI programmed in such a way that they can work together under a single command. These cobots can be used in packing, sealing, filling, and moving pharmaceutical products. Augmented reality technology can be used in the safety and packing of products, R&D, quality assurance, and customer experience.53–55 A digital twin refers to a digital clone of the physical process, like an activity performed by a machine to understand and optimize its performance. It combines empirical data and mechanistic stimulated models to produce highly efficient models with real-time data. Digital twins enable humans to understand the impact of deviation on the performance of any operation or process.56,57
COMPUTATIONAL FLUID DYNAMICS: THE SMART WAY TO TACKLE PHARMACEUTICAL PROBLEMS
Technology Vision 2020 is a document that produces futuristic ideas for industries. Computational Fluid Dynamics (CFD) technology has been a part of that document. This technology is mainly used to examine the fluid flow and heat transfer parameters.58 It works on the principle of the law of conservation of mass, energy, and momentum. The technologies have been used in various sectors; for example, CFD calculates and drag phenomena in aerodynamics. CFD analysis follows three main steps, pre-processing, solution, and post-processing.59
Pre-Processing – the analysis takes place in four different steps. The first is identifying the flow region, the second is the geometrical representation of the flow region, the third is defining an appropriate mesh for that region, and finally, the application of fluid dynamics principles. The software follows all the steps, and finally, the software will design an equation that needs to be solved in the next step.
Solution – once the problem is defined as an equation, this process uses a trial-and-error strategy to compute the solution. All these have been computerized.
Post-Processing – the results are being analyzed in the Post-processing step. The solution provides the entire data of the problem, represented on the flow field, created by plotting flow variables on the 3D region of interest. The analyst analyzes these plots.
CFD technology is very well applied in pharmaceutical industries. It has been a vital tool for analyzing various process variables like mixing, separation, fluid transfer, etc. The CFD software is used to develop stimulation for fluid flow analysis, chemical transport, and heat transfer.60
APPLICATIONS OF COMPUTATIONAL FLUID DYNAMICS IN PHARMACEUTICAL PROCESS
CFD in Mixing Process- the mixing equipment commonly used in pharmaceutical industries are static mixers, homogenizers, stirred tanks, and emulsifiers. Static mixers are mainly applicable for the mixing of high viscous fluids. CFD is used in static mixers to analyze their efficiency and mixing capacity. Stirred mixers are large mixing tanks that primarily produce proper mixing and stirring of contents. These are available in various sizes and types of impellers. CFD methods are used to analyze the vessel’s flow characteristics and predict the impeller’s influence on mixing. It can also determine the shear stress distribution within the stirred mixers. The evaluation of shear stress distribution is essential because it describes the dissolution, dispersion, and emulsification properties.61
CFD in Separation Process – precipitation, centrifugation, and crystallization are used as separation techniques in pharmaceutical industries. Centrifugation techniques at an industrial scale are generally used for thickening, separating solid from liquid, and post-treatment. CFD methods can study the centrifuge design and modify the plan if necessary. The CFD technique also analyzes the performance of separation devices like cyclones and scrubbers. For example, the CFD method was used to analyze the separation efficiency of cyclone devices for particle size, and about 90% of 10-µm sized particles were separated. Still, only 10% of 10-µm sized particles were able to get separated.62
CFD in Drying Process – CFD technology is used to analyze the performance of spray dryers at an industrial scale before drying or making any significant changes in the designs of dryers. It is also used to analyze the congregation changes during the drying process. It helps avoid unnecessary money loss and minimize risk.
CFD in Packing Process – especially in the case of liquid formulations like syrups, suspensions, and emulsions packed in bottles. If the filing of products gets delayed can lead to a severe market issue; other problems during filing like spilling, splashing, and frothing can also lead to packing problems. To overcome such issues, CFD is carried out over the filing lines to analyze any disruption present filing lines before making any significant changes in the geometry of filing lines. CFD is used to optimize the unit operations in pharmaceutical industries during manufacturing. CFD technologies are also used in designing drug delivery systems.
CFD in Designing of Inhalers – Inhalers are mainly used to treat severe lung diseases like asthma, COPD, emphysema, and cystic fibrosis. Metered Dose Inhalers (MDI) and dry powder inhalers are the most common inhalers. By using CFD modeling techniques, pharmaceutical industries started to modify and optimize inhaler designs to make them easy to use and improve the efficiency of inhalers and the reproducibility of the drug product. The model simulation results were used to determine high-velocity regions, shear forces, circulation loops, and particle residence time for larger and smaller particles. The trajectories of drug particles inside the lung can be traced using CFD technology.62
CONCLUSION WITH FUTURE PERSPECTIVES
In this digitalized world, CADD has given a whole new insight into the pharmaceutical industry and drug discovery process innovations and development. Computational technologies and advanced software’s helped the pharmaceutical industries overcome significant problems faced in drug discovery, i.e., longer time issues and higher costs for developing new drug molecules. The concept of computers is constantly growing, with new and advanced technologies like artificial intelligence and machine learning. AI and ML have immense potential to solve complex problems and build innovative technologies, making an automated and digitalized world. The idea of automation and robotics has also positively impacted pharmaceutical sectors. It has unleashed the potential of researchers to innovate new ideas to solve complex problems in the drug discovery process and altogether changed the era of pharmaceutical industries from man to no man industries, i.e., most pharmaceutical industries are now shifting towards machines and robotics, making a complete automated system as discussed in this article. But still, a lot more research and innovation are to be done in the field of pharmaceutical industries so that every sector can develop and produce the most innovative and effective medicines in the market. It can be possible only through artificial intelligence and machine learning. By looking at the present scenario, the pharmaceutical industry and drug discovery process have a bright future because I believe that the concept of AI is the ultimate future.
References
- Kaplan A, Haenlein M. Siri, Siri, in my hand: Who’s the fairest in the land? On the interpretations, illustrations, and implications of artificial intelligence. Bus Horiz. 2019;62(1):15-25. [CrossRef] | [Google Scholar]
- Selvaraj C, Chandra I, Singh SK. Artificial intelligence and machine learning approaches for drug design: challenges and opportunities for the pharmaceutical industries. Mol Divers. 2022;26(3):1893-913. [PubMed] | [CrossRef] | [Google Scholar]
- Henry J, Wlodkowic D. Towards high-throughput chemobehavioural phenomics in neuropsychiatric drug discovery. Mar Drugs. 2019;17(6):340 [PubMed] | [CrossRef] | [Google Scholar]
- Ghosh S, Mitra I. [Jun 15 2022];Artificial Intelligence and Robotics Leveraging artificial intelligence and robotics for sustainable growth [internet]. Array. 2017 [PubMed] | [CrossRef] | [Google Scholar]
- Mijwel MM. Parkinson’s Disease Foundation History of Artificial Intelligence. 2015 [Jul 03 2022]. History of Artificial Intelligence. Research Gate.
- Haenlein M, Kaplan A. A brief history of artificial intelligence: On the past, present, and future of artificial intelligence. Calif Manag Rev. 2019;00:1-10. [PubMed] | [CrossRef] | [Google Scholar]
- Turing A. Computing machinery and intelligence. Mind. 1950;59(236):433-60. [PubMed] | [CrossRef] | [Google Scholar]
- Haenlein M, Kaplan A. A brief history of artificial intelligence: On the past, present, and future of artificial intelligence. Calif Manag Rev. 2019;61(4):1-10. [PubMed] | [CrossRef] | [Google Scholar]
- Weizenbaum J. ELIZA software, developed in. 1966
- Campbell M, Hoane AJ, Hsiung Hsu F. Deep blue. Artif Intell. 2002;134(1-2):57-83. [CrossRef] | [Google Scholar]
- [CrossRef] | [Google Scholar]
- Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, et al. Mastering the game of go with deep neural networks and tree search. Nature. 2016;529:484-9. [PubMed] | [CrossRef] | [Google Scholar]
- Mijwel MM. Parkinson’s Disease Foundation History of Artificial Intelligence. 2015 [Jul 03 2022]. History of Artificial Intelligence, Researchgate, Research Gate.
- Zador AM. A critique of pure learning and what artificial neural networks can learn from animal brains. Nat Commun. 2019;10(1):3770 [PubMed] | [CrossRef] | [Google Scholar]
- Alzahab NA, Apollonio L, Di Iorio A, Alshalak M, Iarlori S, Ferracuti F, et al. Hybrid Deep Learning (HDL)-based Brain-Computer Interface (BCI) systems: a systematic review. Brain Sci. 2021;11(1):75 [PubMed] | [CrossRef] | [Google Scholar]
- Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: methods for virtual ligand screening and profling. Br J Pharmacol. 2007;152(1):9-20. [PubMed] | [CrossRef] | [Google Scholar]
- Ahmed Z, Mohamed K, Zeeshan S. Artifcial intelligence with multi-functional machine learning platform development for better healthcare and precision medicine. Database (Oxf). 2020;2020(10):1-35. [PubMed] | [CrossRef] | [Google Scholar]
- Sakellaropoulos T, Vougas K, Narang S, Koinis F, Kotsinas A, Polyzos A, et al. A deep learning framework for predicting response to therapy in cancer. Cell Rep. 2019;29(11):3367-73.e4. [PubMed] | [CrossRef] | [Google Scholar]
- Poggio T, Banburski A, Liao Q. Theoretical issues in deep networks. Proc Natl Acad Sci U S A. 2020;117(48):30039-45. [PubMed] | [CrossRef] | [Google Scholar]
- Del Fiol G, Michelson M, Iorio A. A deep learning method to automatically identify reports of scientifically rigorous clinical research from the biomedical literature: comparative analytic study. 2018;20(6):10281 [PubMed] | [CrossRef] | [Google Scholar]
- Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging. 2018;9(4):611-29. [PubMed] | [CrossRef] | [Google Scholar]
- Sharma P. What are Graph Neural Networks, and how do they work, Data Science Blogathon. 2022 [Jun 25 2022].
- Sharma P. What are Graph Neural Networks, and how do they work, Data Science Blogathon. 2022 [Jun 25 2022].
- Trabelsi A, Chaabane M, Ben-Hur A. Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics. 2019;35(14):269-77. [PubMed] | [CrossRef] | [Google Scholar]
- Ben-Bassat I, Chor B, Orenstein Y. A deep neural network approach for learning intrinsic protein-RNA binding preferences. Bioinformatics. 2018;34(17):638-46. [PubMed] | [CrossRef] | [Google Scholar]
- Graupe D, Vern B33, Vern B. On the interrelations between artificial and physiological neural networks. Neurol Res. 2001;23(5):482-8. [PubMed] | [CrossRef] | [Google Scholar]
- Lee D, Yoon SN. Application of artificial intelligence-based technologies in the healthcare industry: opportunities and challenges. Int J Environ Res Public Health. 2021;18(1):271 [PubMed] | [CrossRef] | [Google Scholar]
- Volk MJ, Lourentzou I, Mishra S, Vo LT, Zhai C, Zhao H, et al. Biosystems design by machine learning. ACS Synth Biol. 2020;9(7):1514-33. [PubMed] | [CrossRef] | [Google Scholar]
- Jeon J, Nim S, Teyra J, Datti A, Wrana JL, Sidhu SS, et al. A systematic approach to identify novel cancer drug targets using machine learning, inhibitor design and high-throughput screening. Genome Med. 2014;6(7):57 [PubMed] | [CrossRef] | [Google Scholar]
- Mamoshina P, Volosnikova M, Ozerov IV. Machine learning on human muscle transcriptomic data for biomarker discovery and tissue-specific drug target identifcation. Front Genet. 2018;12(9):242 [PubMed] | [CrossRef] | [Google Scholar]
- Huang S, Cai N, Pacheco PP, Narrandes S, Wang Y, Xu W, et al. Applications of Support Vector Machine (SVM) learning in cancer genomics. Cancer Genomics Proteomics. 2018;15(1):41-51. [PubMed] | [CrossRef] | [Google Scholar]
- Yang X, Wang Y, Byrne R, Byrne R, Schneider G, Yang S, et al. Concepts of artificial intelligence for computer-assisted drug discovery. Chem Rev. 2019;119(18):10520-94. [PubMed] | [CrossRef] | [Google Scholar]
- Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, et al. A guide to deep learning in healthcare. Nat Med. 2019;25(1):24-9. [PubMed] | [CrossRef] | [Google Scholar]
- Ramsundar B, Liu B, Wu Z, Verras A, Tudor M, Sheridan RP, et al. Is Multi-task deep learning practical for pharma?. J Chem Inf Model. 2017;57(8):2068-76. [PubMed] | [CrossRef] | [Google Scholar]
- Mayr A, Klambauer G, Unterthiner T, Hochreiter S. DeepTox: toxicity prediction using deep learning. Front Environ Sci. 2016;3(80):1-15. [CrossRef] | [Google Scholar]
- Rao VS, Srinivas K, Sujini GN. Protein-protein interaction detection: methods and analysis. Int J Proteomics. 2014;2014:1-12. [CrossRef] | [Google Scholar]
- Ding Z, Kihara D. Computational methods for predicting protein-protein interactions using various protein features. Curr Protoc Protein Sci. 2018;93(1):62 [PubMed] | [CrossRef] | [Google Scholar]
- Wang S, Sun S, Li Z. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLOS Comput Biol. 2017;13(1):1005324 [PubMed] | [CrossRef] | [Google Scholar]
- Dobchev D, Karelson M48 M.. Have artificial neural networks met expectations in drug discovery as implemented in QSAR framework?. Expert Opin Drug Discov. 2016;11(1):627-39. [PubMed] | [CrossRef] | [Google Scholar]
- Hong H, Rua D, Sakkiah S, Selvaraj C, Ge W, Tong W, et al. Consensus Modeling for Prediction of Estrogenic Activity of Ingredients Commonly Used in Sunscreen Products. Int J Environ Res Public Health. 2016;13(10):958 [PubMed] | [CrossRef] | [Google Scholar]
- Meftahi N, Walker ML, Enciso M, Smith BJ. Predicting the enthalpy and Gibbs energy of sublimation by QSPR modeling. Sci Rep. 2018;8(1):9779 [PubMed] | [CrossRef] | [Google Scholar]
- Hefti FF. Requirements for a lead compound to become a clinical candidate. BMC Neurosci. 2008;9(8-S3):7 [PubMed] | [CrossRef] | [Google Scholar]
- Xu Y, Pei J, Lai L. Deep learning based regression and multiclass models for acute oral toxicity prediction with automatic chemical feature extraction. J Chem Inf Model. 2017;57(11):2672-85. [PubMed] | [CrossRef] | [Google Scholar]
- Li X, Xu Y, Lai L, Pei J. Prediction of human cytochrome P450 inhibition using a multi-task deep auto-encoder neural network. Mol Pharm. 2018;15(10):4336-45. [PubMed] | [CrossRef] | [Google Scholar]
- Avdeef A, Box KJ, Comer JE, Hibbert C, Tam KY. PH-metric logP 10. Determination of liposomal membrane-water partition coefficients of ionizable drugs. Pharm Res. 1998;15(2):209-15. [PubMed] | [CrossRef] | [Google Scholar]
- [PubMed] | [CrossRef] | [Google Scholar]
- Montague G, Morris J. Neural-network contributions in biotechnology. Trends Biotechnol. 1994;12(8):312-24. [PubMed] | [CrossRef] | [Google Scholar]
- [PubMed] | [CrossRef] | [Google Scholar]
- [PubMed] | [CrossRef] | [Google Scholar]
- Marcus Ehrhardt P.B.. Available from. Strategy and [internet]. 2016 Digitization in Pharma Gaining an edge in operations.
- [PubMed] | [CrossRef] | [Google Scholar]
- Arden NS, Fisher AC, Tyner K, Yu LX, Lee SL, Kopcha M, et al. Industry 4.0 for pharmaceutical manufacturing: preparing for the smart factories of the future. Int J Pharm. 2021;602:120554 [PubMed] | [CrossRef] | [Google Scholar]
- [PubMed] | [CrossRef] | [Google Scholar]
- Stracquatanio, A.. Harnessing Augmented Real Pharm Manuf Pharm Online. 2018 [PubMed] | [CrossRef] | [Google Scholar]
- [PubMed] | [CrossRef] | [Google Scholar]
- [PubMed] | [CrossRef] | [Google Scholar]
- . GE Res Digit Twin. 2020 [PubMed] | [CrossRef] | [Google Scholar]
- Weiner S.C.. Technology Vision 2020. The US chemical industry. American Chemical Society. 1998;72:915-21. [PubMed] | [CrossRef] | [Google Scholar]
- Pordal HS, Matice CJ, Fry TJ. The role of computational fluid dynamics in the pharmaceutical industry. Pharm Technol. 2002;26(2):1-8. [PubMed] | [CrossRef] | [Google Scholar]
- Sharma T, Mankoo A, Sood V. Artificial intelligence in advanced pharmacy. Int J Sci Res Arch. 2021;6(2):47-54. [PubMed] | [CrossRef] | [Google Scholar]
- [PubMed] | [CrossRef] | [Google Scholar]
- [PubMed] | [CrossRef] | [Google Scholar]
